High-Speed Holography of Cells Spots Physical Beacons of Disease
By Ken Kingery
Measuring the variability of thickness within thousands of cells per minute spots physical signs of cancer and carcinogenic exposure

Biomedical engineers at Duke University have engineered a holographic system capable of imaging and analyzing tens of thousands of cells per minute to both discover and recognize signs of disease.
In the proof-of-concept demonstration, the technique distinguished between healthy samples and either cancerous or carcinogen-exposed, pre-cancerous cells with nearly 100% accuracy, using just four basic cellular physical parameters out of a holographic panel of 25. The results point toward a promising screening or diagnostic technology that is simpler and cheaper to use than current standard practices, making it a potential target for use in remote, low-resource settings.
The research appears online on November 30 in the journal Frontiers in Physics.
“The cells are flying through the scanner so fast that if the computer didn’t slow them down on the screen, you wouldn’t even be able to see them,” said Adam Wax, professor of biomedical engineering at Duke. “We were very excited to be able to image this many cells at once because it points toward this technology’s potential for point-of-care diagnostics.”
In a traditional cellular diagnostic process, cells obtained from tissue scraping or fine needle biopsies are put on a slide and stained for a trained physician to carefully magnify and inspect. But in some cases, the staining process can damage the cells, and getting the slides to the eyes of trained experts can take days if not weeks.
In the new holographic imaging approach, sample cells could be rinsed off of the collection instrument into a biocompatible solution and inserted into a microfluidic chip. The small device diverts the sample into a series of parallel channels that pass beneath a line camera, kind of like a conveyor belt passing newly manufactured items beneath an autonomous scanner for inspection. When slowed down, the march of cell images is reminiscent of green characters cascading down a computer screen in The Matrix.
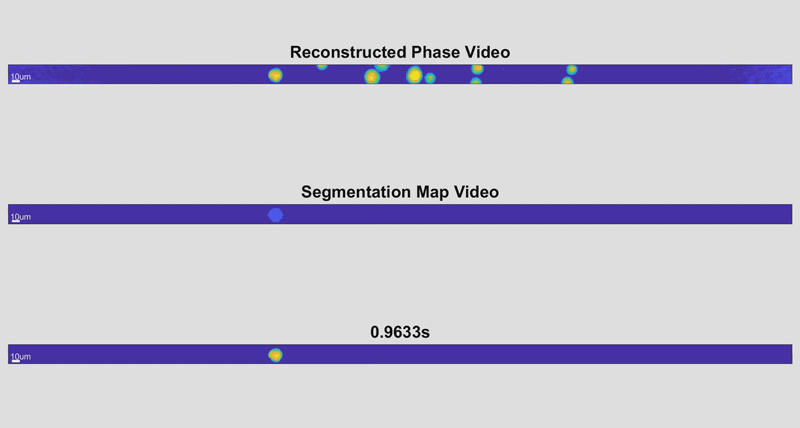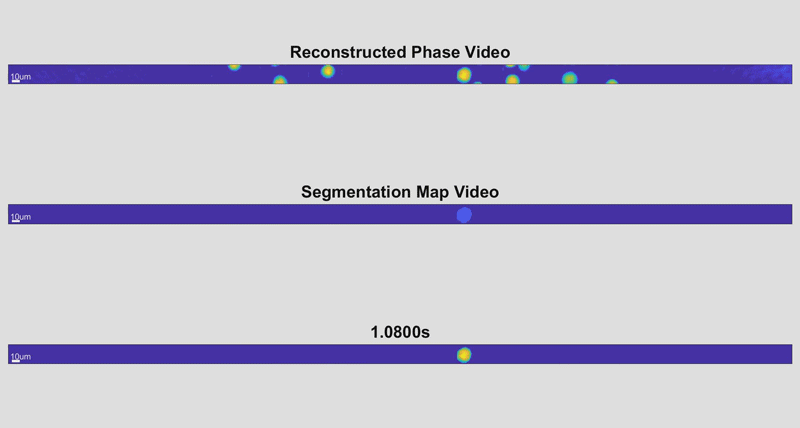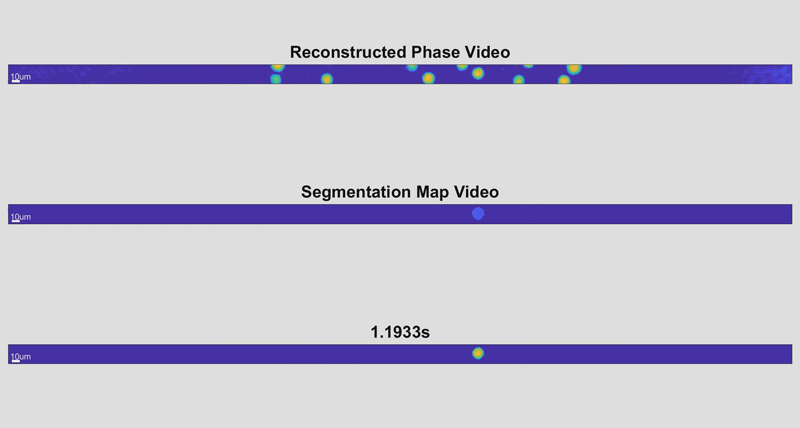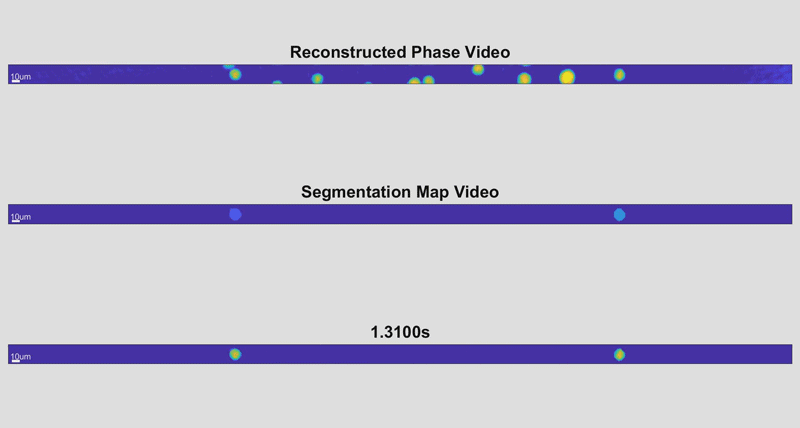 As the cells fly by, the device automatically picks out whole cells and shines a light through them. Based on the difference in time it takes for the light to return from its journey and an unimpeded reference beam, the camera can determine the topography of each cell and calculate features such as its shape and height. While these may sound like simple measurements, when they are combined with tens of thousands of data points and advanced deep learning techniques, they can become beacons for spotting disease.
As the cells fly by, the device automatically picks out whole cells and shines a light through them. Based on the difference in time it takes for the light to return from its journey and an unimpeded reference beam, the camera can determine the topography of each cell and calculate features such as its shape and height. While these may sound like simple measurements, when they are combined with tens of thousands of data points and advanced deep learning techniques, they can become beacons for spotting disease.
“It takes about 30 seconds to process a 1-milliliter sample that could contain more than a half-million cells,” said Cindy Chen, a doctoral student working in Wax’s lab and first author of the paper. “And once it’s finished, a pathologist could pull up individual data for any of the cells that were imaged for closer inspection.”
To demonstrate the potential of this approach, Wax and Chen used it to image a combined 8500 cells from three different cell lines—healthy breast cells, cancerous breast cells and pre-cancerous breast cells that had been exposed to cadmium, a known potent carcinogen. While the technology captures 25 separate physical characteristics of the cells, the researchers discovered that in this case, only four were necessary for differentiating the three types.
Using a machine learning algorithm, the device was able to successfully categorize each cell with 98% to 99% accuracy using just the 2D area it occupies, the 3D space it takes up, its shape and its average height.
“This is a simple, label-free imaging technique that is getting data similar to that of flow cytometry, which looks at cells one at a time and typically requires fluorescent markers to be introduced,” said Chen. “The idea is that, by having this wealth of quantitative data, you can separate the cells better than if you were using one single metric.”
“We’re now trying to figure out how to put this technology at the point-of-care to figure out if people have been exposed to these carcinogens in the developing world, like maybe arsenic in the ground water,” added Wax. “For example, you might brush some cells off of the top of someone’s mouth to see if they’re being exposed to carcinogens in their drinking water.”
This research was supported by the National Institutes of Health (1R21ES029791).
CITATION: “Automated Classification of Breast Cancer Cells Using High-Throughput Holographic Cytometry,” Cindy X. Chen, Han Sang Park, Hillel Price and Adam Wax. Frontiers in Physics, Nov. 30, 2021. DOI: 10.3389/fphy.2021.759142
