Proteins Use Force to Make New Connections
Michaela Martinez
New imaging approach visualizes how applying force to proteins alters complex formations
Most researchers today understand biology through the principles of biochemistry. Cells communicate and activate processes via chemical signals, and traditional medicine has long focused on how to treat disease by modifying those signals.
This approach, however, ignores a major factor that affects cell behavior: physical mechanics.
“Applying physical force to a cell can alter and control the cell’s structure and behavior,” explains Brenton Hoffman, the James L. and Elizabeth M. Vincent Associate Professor of Biomedical Engineering at Duke University. “For example, forces generated by flowing blood helps veins and arteries grow into the correct shape and behave normally.”
But altered mechanics can also cause abnormal behaviors seen in diseases such as breast cancer.
“A breast tumor is normally discovered when someone feels a stiff lump, which causes cells to generate more mechanical force that can lead to cellular migration,” says Hoffman. “Recent work has indicated that these physical changes could actually be a trigger for cancer to spread.”
Because the field of mechanobiology is comparatively new, researchers are still developing tools that tackle basic questions regarding how different mechanical cues can disturb cells and their signaling pathways. In a new paper that appeared in Developmental Cell in March, Hoffman and his team described a new imaging approach to visualize how forces can affect a protein and its surrounding environment in a living cell.
Before this work, there were two primary ways to study the effects of mechanical forces on protein behavior. The first way involves applying forces to individual proteins, or protein pairs, that have been purified and removed from the cell. While this can precisely determine if force affects the protein behavior, it can’t probe how these behaviors affect other proteins or cellular structure.
The other approach involves catching proteins that leave cellular structures when these structures are forced to remain still. While this approach can identify a large number of proteins that have some force-sensitivity, it does not tell researchers much about how these proteins are interacting with each other and forming complex structures.
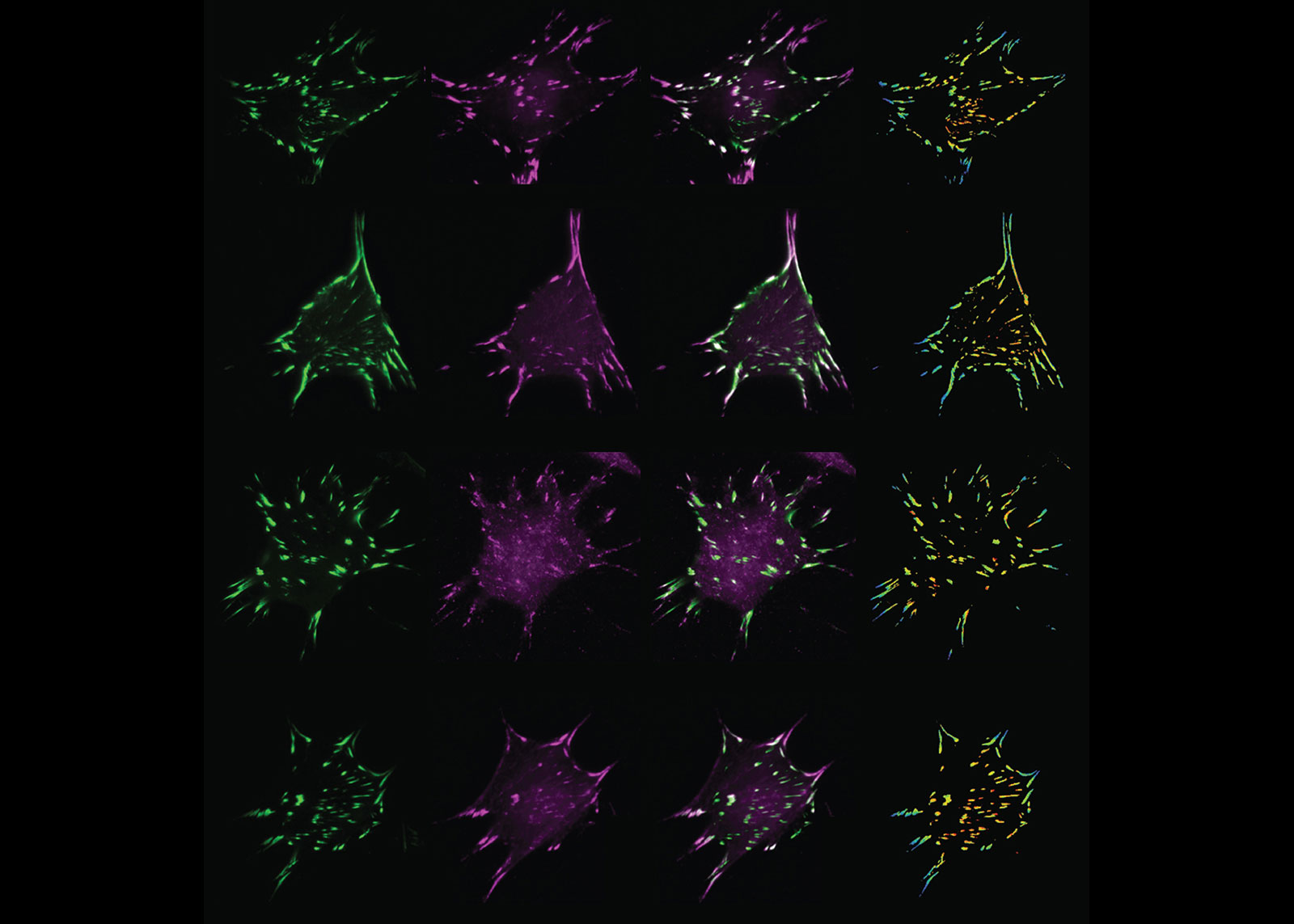
To bridge the gap between these approaches, Hoffman and his team developed FTC, or fluorescence-tension co-localization. FTC enables researchers to study how local concentrations of a protein changes when a different protein gets physically poked.
“FTC allows us, for the first time, to see how mechanical forces experienced by one protein affect its ability to attract other proteins,” says Hoffman. “This is a major accomplishment because altering protein proximity is one of the main ways that signaling pathways get activated, which controls cell behavior.”
As a proof of concept, Hoffman and his team studied the linker protein vinculin, which is found in sub-cellular structures known as focal adhesions that provide physical support to cells. By imaging and analyzing over 5,000 cells and over 500,000 focal adhesions, the team determined how 20 other proteins moved in response to changes in the forces experienced by vinculin.
While they observed that a large majority of the proteins moved toward vinculin when force was applied, the team also observed several additional factors that influenced which proteins were recruited by vinculin, like the amount of force applied, where the force was applied, and the maturity of the focal adhesion containing the vinculin.
For example, if the vinculin was part of an immature focal adhesion, tension caused increased interactions with proteins that regulate adhesion to the surrounding support structure. In contrast, if vinculin was part of a mature focal adhesion, tension increased interactions with proteins that regulate the ability of the cell to contract. This context dependence likely underlies the strong, and diverse, effects of mechanical stimuli on cells.
This work, Hoffman says, is foundational to advancing the field of mechanobiology, as identifying these types of force-sensitive protein relationships will help researchers decipher force-sensitive signaling pathways. The ability to understand how the surroundings impact vinculin’s interactions with other proteins could also help researchers shut down the signaling that drives undesirable cell behaviors, or even disease.
“There are a lot of disease states that lack an effective or side-effect-free chemical treatment option, whether it’s cancer, asthma, cardiovascular disease or muscular dystrophy. These all have a clear mechanical component, suggesting an untapped means of identifying novel therapeutic targets,” says Hoffman. “Making the tools necessary to do these basic studies of how force affects the signals that proteins send to their surroundings is key to understanding these mechanical effects, and it has the potential to influence fields from tissue engineering to medicine.”
