Machine Learning Platform Identifies Activated Neurons in Real-Time
Michaela Kane
Streamlined AI immediately and accurately maps activated neurons to help learn how the brain works
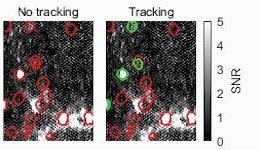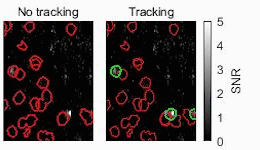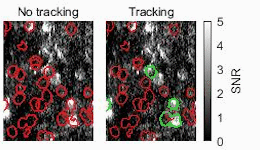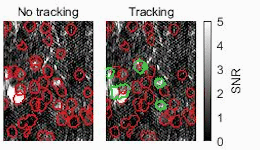

Biomedical engineers at Duke University have developed an automatic process that uses streamlined artificial intelligence (AI) to identify active neurons in videos faster and more accurately than current techniques.
The technology should allow researchers to watch an animal’s brain activity in real time, as they are behaving.
The work appears May 20 in Nature Machine Intelligence.
One of the ways researchers study the activity of neurons in living animals is through a process known as two-photon calcium imaging, which makes active neurons appear as flashes of light. Analyzing these videos, however, typically requires a human circling every burst of intensity they see in a process called segmentation. While this may seem straightforward, these bursts often overlap in spaces where thousands of neurons are imaged simultaneously. Analyzing just a five-minute video this way could take weeks or even months.
“People try to figure out how the brain works by recording the activity of neurons as an animal does a behavior to study the relationship between the two,” said Yiyang Gong, the primary author on the paper. “But manual segmentation creates a big bottleneck and doesn’t allow researchers to see the activation of the neurons in real-time.”
Gong, an assistant professor of biomedical engineering, and Sina Farsiu, a professor of biomedical engineering, previously addressed this bottleneck in a 2019 paper, where they shared the development of a deep-learning platform that maps active neurons as accurately as humans in a fraction of the time. But because videos can be tens of gigabytes, researchers still have to wait hours or days for them to process.
Now, the team is making their platform work in real-time.
“Our goal was to improve our approach to be more intelligent so it can target and learn from the important data in the videos rather than parse through all of the extra noise,” said Gong.
To make their approach more intelligent, the team developed signal processing algorithms that pre-process the data before being analyzed by the neural network. These algorithms help improve the signal-to-noise ratio and remove the background fluctuations in the video, highlighting active neurons while obscuring inactive neurons and other unneeded data.
The team also adapted their neural network to scan fewer layers, because it doesn’t need as much data to learn how to accurately identify and segment the activated neurons.
The result is a system that features an unusual upgrade. Not only is it an order of magnitude faster than their previous work, it’s also slightly more accurate.
Because their platform can highlight active neurons so quickly, researchers can use the tool to detect neurons in real-time and examine how certain activation patterns match animal behavior. Due to the tool’s usefulness in neuroscience experiments, the team has made a version of the network available online. (LINK PLS)
“Rather than wait until the end of an experiment, the speed of our network allows us to learn things during the experiment,” said Gong. “We now have a new potential to explore how different kinds of stimulation can affect neuronal activation and animal behavior.”
The team is already exploring new ways to continue to improve their tool for wider use.
“The algorithm can always use further optimization,” said Gong. “We’ve shown that this works really well for the two-photon calcium imaging, but there are a lot of different optical microscopes in neuroscience, and ultimately we’d like to make a neural network that works for all of these imaging modalities.”
This work was supported by the BRAIN Initiative (NIH 1UF1-NS107678, NSF 3332147), the NIH New Innovator Program (1DP2-NS111505), the Beckman Young Investigator Program, the Sloan Fellowship, and the Vallee Young Investigator Program. The team also acknowledge Zhijing Zhu for early characterization of the SUNS.
CITATION: “Segmentation of Neurons from Fluorescence Calcium Recordings Beyond Real Time,” Yijun Bao, Somayyeh Soltanian-Zadeh, Sina Farsiu, Yiyang Gong. Nature Machine Intelligence, May 20, 2021. DOI: 10.1038/s42256-021-00342-x
