‘Embrace the Ditch,’ and Other Lessons Learned in Duke CEE’s Overture Engineering
Civil and environmental engineering students learn to design buildings within less-than-optimal parameters in a collaborative capstone course
We’re sorry, but that page was not found or has been archived. Please check the spelling of the page address or use the site search.
Still can’t find what you’re looking for? Contact our web team »
Read stories of how we’re teaching students to develop resilience, or check out all our recent news.

Civil and environmental engineering students learn to design buildings within less-than-optimal parameters in a collaborative capstone course

On a Star Wars-themed field of play, student teams deployed small robots they had constructed
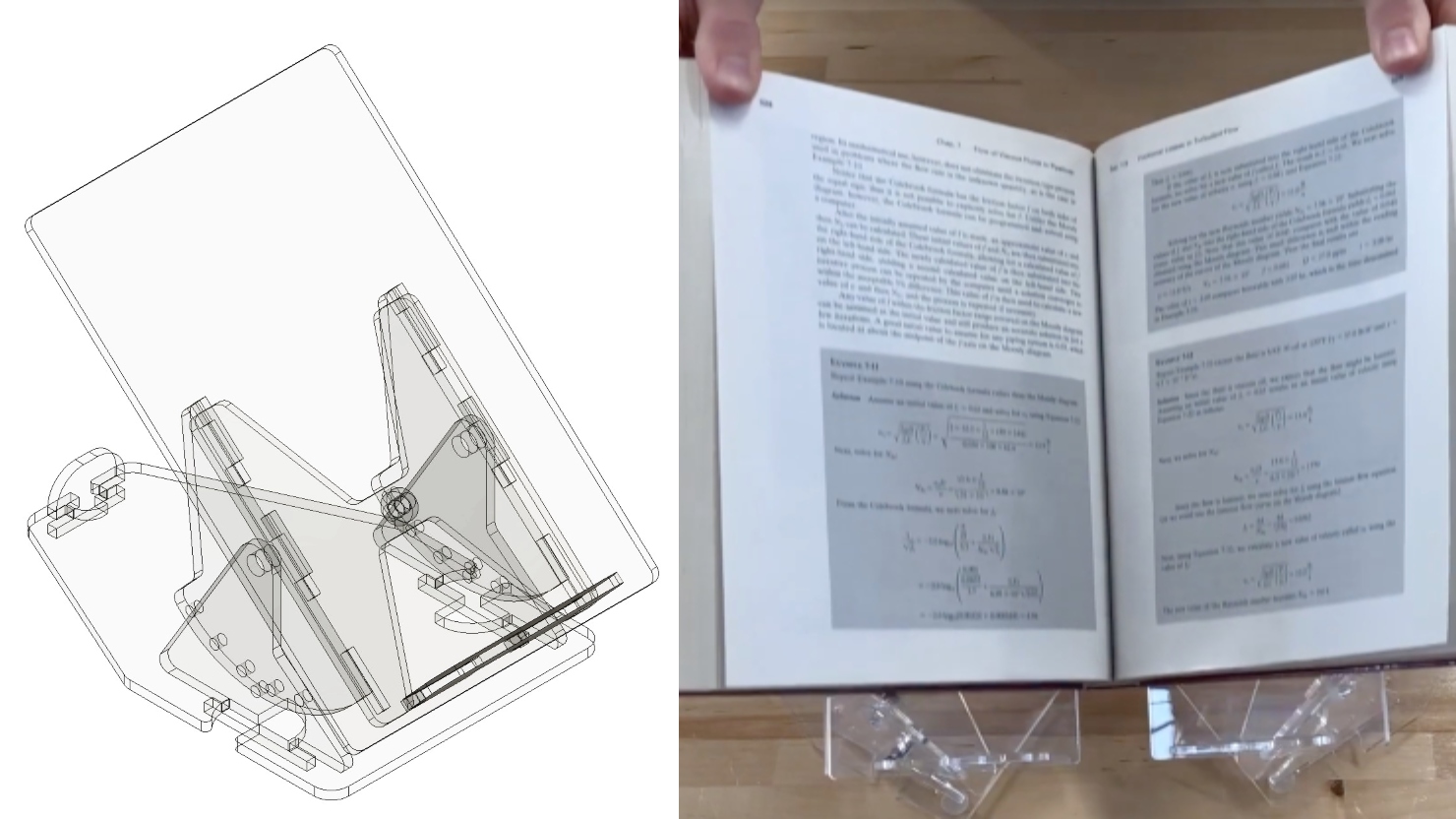
Two projects from First-Year Design course are patent-pending. Student surveys suggest the course also fosters teamwork, leadership and communication skills.
Feb 9
Feb 9
Join us for tea and snacks on the 3rd floor of Gross Hall! This is a great chance to meet colleagues in a casual environment. All Welcome!
1:00 pm – 1:00 pm Gross Hall, Ahmadieh Family Atrium, 3rd Floor
Feb 10
Engineering Master’s students: Attend Career Online Drop-In Hours to connect with a Career Coach for quick questions and feedback on your application documents.
12:00 pm – 12:00 pm Online