Illuminating the Dark Genome
By Ellery Jones
Duke’s Center for Advanced Genomic Technologies shines brightly on the horizon

Each stepping stone in the history of genetics has led us closer to understanding the complex links between our DNA and our everyday lives. From Mendel’s humble pea plant studies in the 19th century to the completion of the monumental Human Genome Project in 2003, scientists have made incredible progress in a short amount of time.
Over the past two decades, researchers’ abilities to sequence, organize and even edit the genome have skyrocketed with the development of rapid sequencing and bioinformatic technologies. Recently, geneticists have discovered a new route on the roadmap that ties our genetic code to our health and well-being, sometimes referred to as the “dark genome.”
These stretches of our genome do not instruct our cells on how to make specific proteins but rather help control when, where and to what extent our protein-coding genes are activated or deactivated. Here at Duke, the Center for Advanced Genomic Technologies (CAGT) is launching the scientific community into the frontier of studying how the dark genome helps control human health and disease.
The CAGT began with conversations between Maria Ciofani, assistant professor of immunology; Gregory E. Crawford, associate professor of pediatrics; Timothy Reddy, associate professor of biostatistics & bioinformatics; Kris Wood, assistant professor of pharmacology & cancer biology; and Charles Gersbach, the Rooney Family Associate Professor of Biomedical Engineering and director of the CAGT.
Gersbach recognized that this core group needed an identity to expand and incorporate other labs and people into it, and from these five founding members, the CAGT was born. In the brief time since its founding, the center has grown to a community of 27 professors across fields of engineering, biology, medicine, bioinformatics and more.

The Duke steering committee. From left to right, Ciofani, Crawford, Gersbach, Reddy, and Wood.
Jeni Reininga-Craven, executive director of the CAGT, has watched the center blossom into an expansive and diverse research coalition from those informal beginnings.
“We recognized that there were really stellar collaborations and big research impacts through these five steering committee members,” she said. “Not only were these groups successful in their research, but in their other impacts, such as providing a rich multidisciplinary training environment, receiving grant funding from diverse agencies and participating in national consortia.”
Taking part in these national consortia, such as the National Institute of Health-backed ENCODE Consortium, the Roadmap Epigenetics Program, and the Somatic Cell Genome Editing Consortium, will allow the CAGT researchers to share their work and connect with others on a worldwide scale.
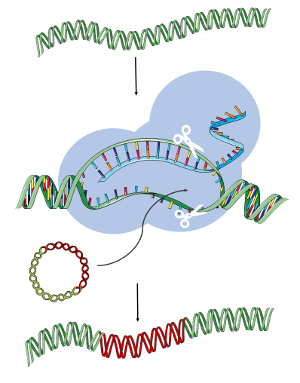
CRISPR technology allows the living genome to be edited with extreme precision, including the insertion of whole genes into target cells.
At this point, one might ask, why choose to study the non-coding aspects of the genome in the context of disease? Aren’t protein-coding genes the stars of the show? According to Reininga-Craven, this is a common misconception. “Limitations in technology are a big reason it hasn’t been studied before,” she explained. “In the last ten years or so, genome-wide association studies have helped us understand the role of the genome in complex diseases. However, researchers didn’t uncover as many variants in protein-coding genes as they thought they would. In fact, about 90 percent of the genetic variation known to exist for complex diseases is undiscovered. That’s why we need to look at the non-coding genome next.”
One of the reasons the CAGT is uniquely suited to tackle significant challenges in genetic research is its multidisciplinary team. “One of the major challenges of genetic research is that genetic data is very difficult to measure and interpret—how do we link regulatory variation of genes to the outcomes of diseases? It’s a complex system,” said ReiningaCraven.
Gersbach, whose lab focuses on the development of Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) genome editing technologies, wholeheartedly agrees. “The primary goal of the CAGT is to unify researchers across the Schools of Engineering, Medicine, and Arts & Sciences to tackle grand challenges of genomics that can’t be addressed by any one approach or discipline,” he said. “One of the challenges is that engineers continue to develop more advanced high-throughput technologies that generate greater and greater amounts of data, and this necessitates innovative bioinformatic approaches to analyze these datasets in robust and statistically sound ways.”
Gersbach offered a concrete and nearly mind-boggling example of the data analysis and interpretation challenges that advanced sequencing technology produces. “For example,” he said, “how do we assign a function to every base pair of the 3 billion base pairs of the human genome, within each of the billions of individual cells in a tissue, across thousands of different people in a patient cohort?”
The answer lies in cooperation between multiple fields of expertise in bioinformatics, statistics and engineering. Collaborations between the CAGT and the Center for Statistical Genetics and Genomics, led by Andrew Allen, professor of biostatistics and bioinformatics, seek to address some of these challenges.
“We all meet together multiple times each week to tackle various projects in these areas,” said Allen. “Several of our faculty, such as Professors Tim Reddy, Raluca Gordan and Bill Majoros, are members of both centers, leading to natural integration across disciplines and research projects.”

Two students work under a hood in the CAGT wet lab at Duke.
The CAGT is poised to make an international impact through its groundbreaking research, but it is dedicated to providing local educational programs and professional development opportunities in its home community as well. Gersbach shared his excitement for the translational potential of the CAGT’s research.
“For the last five or six years,” Gersbach said, “we’ve been developing exciting new technologies, applying them to various disease areas and capitalizing on novel therapeutic directions. The rapid development of genome-based therapeutics is aided by partnerships between medicine and engineering and has led to spinning our technology into patents and the biotechnology industry.”
Reininga-Craven echoed the success of the CAGT’s technology, sharing that some of their recent work has led to the creation of start-up companies and philanthropic pursuits. She also emphasized that one of the major goals of the center is to provide enrichment and professional development opportunities to students, from symposia including renowned leaders in the field to assistance with science communications.
“We want to make sure we have programming that meets the needs of everyone across the spectrum,” said Reininga-Craven. “Let’s build around the people that are here and leverage their unique resources and experiences to aid in peer mentoring and building a community.” Excited to hear ideas from incoming and current students on how the CAGT could provide opportunities for them to grow as scientists, leaders and mentors, she added, “Now’s a great time to hear from trainees—we’re brainstorming!”
Finally, Reininga-Craven shared her vision of the CAGT’s bright future. “In establishing the center,” she said, “we’re also establishing Duke as a thought leader in genomic technology. I think we have the right people, and Duke is the right place, to make major progress.”
Ellery Jones is pursuing her PhD in biomedical engineering and uses tissue-engineered blood vessels to study the development of cardiovascular disease.
