Chilkoti Receives $13.7 Million in Funding to Prototype a Rapid, Portable Diagnostic for Soldiers and Global Health
Ashutosh Chilkoti will explore how to use his lab’s signature diagnostic platform to detect bioterrorism agents and to diagnose breast cancer
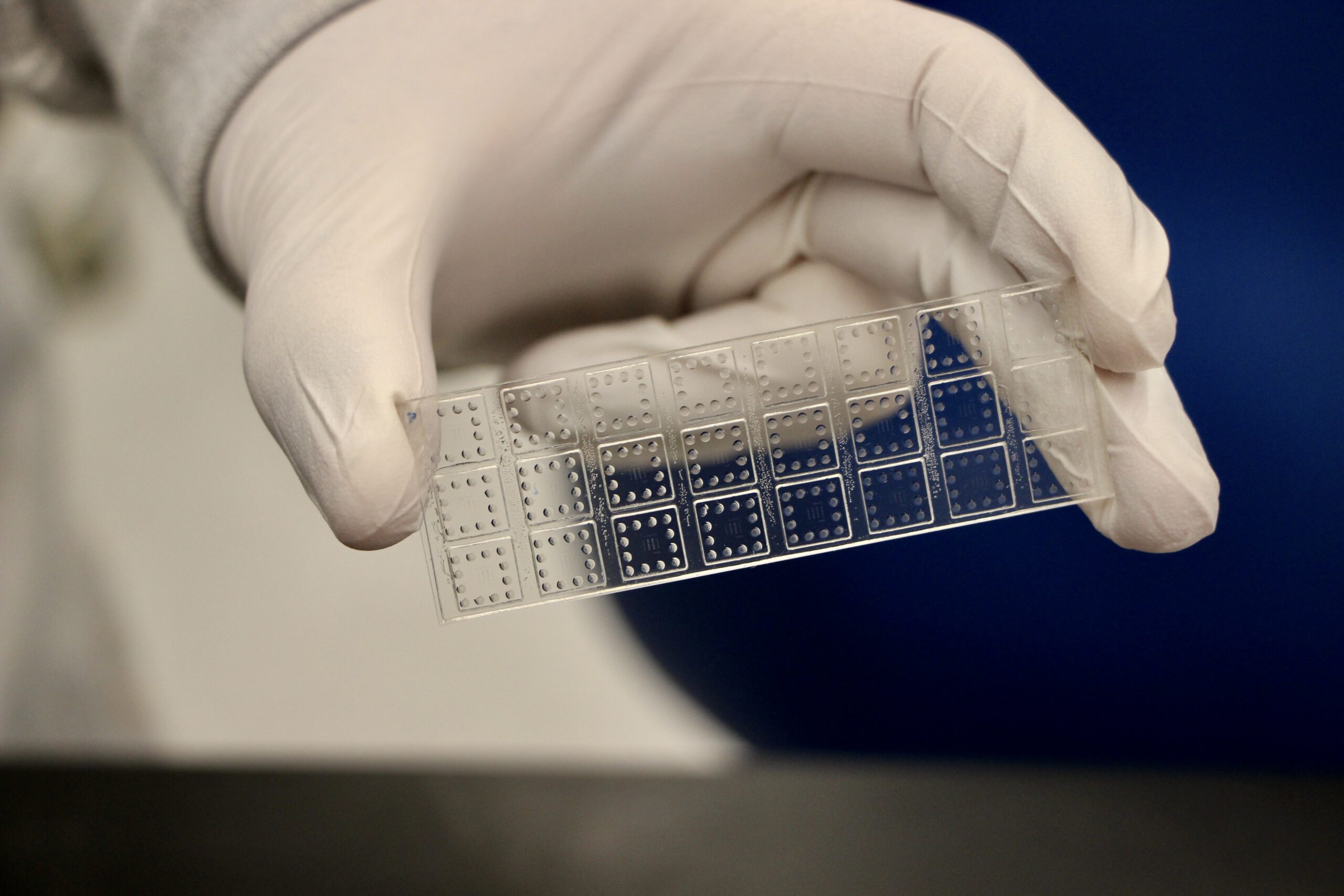
Ashutosh Chilkoti, the Alan L. Kaganov Distinguished Professor of Biomedical Engineering at Duke University, has been awarded two grants totaling $13.7 million in funding to develop a rapid, portable diagnostic test that can detect a variety of illnesses and pathogens.
Both grants involve using the Chilkoti lab’s signature diagnostic platform––the D4 point-of-care test (POCT). Made on a glass slide, the test looks for proteins created from a virus or other infection in blood, serum or saliva samples. Two types of antibodies are printed on the slide’s surface––capture antibodies and detection antibodies, which are tagged with a fluorescent marker. When a sample is placed on the slide, the detection antibodies separate from the array and bind to the target proteins. These antibody-protein pairs then attach to the capture antibodies on the slide, which can be examined using a handheld scanner.
According to Chilkoti, one of the distinguishing features of the platform is the novel polymer brush coating, which prevents non-target proteins from attaching to the slide’s surface.
“By preventing unwanted proteins from binding to the assay the polymer brush makes it easier to detect low levels of target proteins by getting rid of the ‘background noise’ on the chip,” said Chilkoti. “The polymer brush also allows us to store all of the reagents we need on the chip while maintaining a simple design that keeps it portable and easy-to-use.”
In the first grant, which is supported by the Medical CBRN Defense Consortium (MCDC) that provides $10,736,110 in funding over four years, Chilkoti will develop a D4 POCT to detect various toxins, viruses, bacteria and other biowarfare agents in under 30 minutes.
The team will begin by optimizing single biomarkers for four biowarfare agents: ovalbumin, MS2, Pantoea agglomerans and Bacillus globigii spores. After the team has found a successful antibody pair for each target, they’ll continue to optimize the tests until they can get a successful result with a 30 minute incubation time. Once the individual tests are optimized, the team will combine the biomarkers into a single 4-marker array.
As the team optimizes the tool they’ll also work with BioStealth, a commercial subcontractor, to redesign the chip’s format so it can contain and isolate fluids from both biological and environmental samples, allowing for simultaneous testing.
“MCDC is interested in a single point-of-care test for biowarfare agents that can be read within 30 minutes with a hand-held detector”, said Chilkoti. “In the first phase, we will work with simulants, or mimics, of biowarfare agents. Assuming that the results from this simulant panel meet the criteria laid out by MCDC, we will then move into the next phase of this project and develop tests for actual biowarfare related toxins, viruses and bacteria.”
In a second project, Chilkoti and his team will receive $2,990,794 from the National Cancer Institute to diagnose breast cancer in low-resource settings.
According to the World Health Organization, breast cancer is the most common cancer among women, with more than 1.6 million new cases diagnosed each year and more than 500,000 deaths annually. Half of these cases are reported in resource-limited settings, where the majority of deaths from breast cancer occur.
When patients are initially diagnosed with breast cancer, a pathologist must confirm the diagnosis by imaging cells from breast tumors. They also need to identify specific tumor biomarkers, namely the estrogen receptor (ER), the progesterone receptor (PR), human epidermal growth factor receptor-2 (HER2) and the proliferation maker Ki67. Assessing the levels of these biomarkers is essential for determining prognosis and selecting the most effective therapies. However access to both trained pathologists and the necessary diagnostic tools is limited in low-resource settings.
The Chilkoti lab aims to address this discrepancy by using the D4 platform to analyze the cellular and molecular features of breast cancer samples. This new prototype, called the EpiView-D4 point-of-care test, will allow smartphone-based cellular imaging, making it easier for non-pathologists to identify abnormalities in tumor samples. Once malignancy is identified, the device will then quantify ER/PR/HER2/Ki67 levels in the samples in under 30 minutes.
Daniel Joh—currently a plastic surgery resident at Duke Hospital and postdoc in the lab—launched this project in his prior life as a MD/PhD student working in the lab with Chilkoti as part of his PhD.
“In many ways this is Daniel’s brainchild,” said Chilkoti. “He and Jayanta Bhattacharyya, a former postdoc, started the project and collected much of the preliminary data that went into the proposal. Daniel then played an indispensable role in drafting a complex and ambitious proposal that involved my group from BME, Guillermo Sapiro from ECE, Shelley Hwang and her colleagues from DCI, and clinical collaborators in Tanzania. We would not be here today, launching this exciting project, without his efforts.”
The funding will support a multidisciplinary team as they train machine learning algorithms to automatically detect malignant characteristics of breast tissue samples. Once device development is finalized, they’ll conduct pre-clinical validation and clinical investigations of the device, first at Duke University Medical Center, and then at the Kilimanjaro Christian Medical Center in Tanzania to explore how the tool operates in resource-limited settings.
“The impact of this technology lies in its potential to improve breast cancer management worldwide by enabling rapid and accurate diagnosis and subtyping of breast cancers,” said Chilkoti. “This tool could drive timely and appropriate treatment for breast cancer patients and improve the outcomes for hundreds of thousands of women with breast cancer, especially in resource-limited settings.”
